Gueorgui A.Osipov, Dr. of Sci.
|
CURRICULUM VITAE
|
Name: Osipov Gueorgui
Andreevich
Sex: Male
Date of birth: 05 September, 1939
Place of birth: Moscow, Russia
Marital status: widower, adult son and daughter, two grandsons
|
Home address: |
Profsoyusnaya 75-3-38 |
|
|
117342 Moscow |
||
|
Russia |
||
|
Institutional affiliation: |
Doctor of Biology Science |
|
|
Leading Scientist |
||
|
of Academician Yu.Isakov Scientific Group |
||
|
Scientific Center for Heart and Vessels Surgery |
||
|
Russian Academy of Medical Science |
||
|
Institutional Address: |
Sadovo-Kudrinskaya St., 15 |
|
|
Building 2, Room 65 in Filatov Pediatric Hospital |
||
|
101003, Moscow |
||
|
Russia |
||
|
tel. /095/ 254-67-40 |
||
|
fax: /095/ 254-10-39 |
||
|
E-mail: osipovga@rusmedserv.com |
||
Languages: English (fluently) and Russian (native)
Education:
1957-1963 - Moscow Physical and Engineering Institute, Chair of Stable Isotopes.
Degree: M.Sc., Mass Spectrometry. M.Sc. Thesis:” Mass spectrometric ion source with quadripole focusing lenses ”
1963 - 1970 Post-graduate course in Institute of Chemistry and Mechanics, Moscow.
Ph.D. / Candidate of Technical Science/. Thesis: “Thermal decomposition of tetrahydroaluminates of alkaline metals”
1996 - Ph.D./Doctor of Biology Science, microbiology/ Moscow Medical Academy, Chair of Microbiology. Thesis: “Investigation of micro-organisms and microbial communities by using Gas Chromatography - Mass Spectrometry”
Experience:
1963-1978 - Institute of Chemistry and Mechanics, Moscow. Senior Scientist and lead of group of Scientific Instruments /Mass Spectrometry, Nuclear Magnetic Resonance, Electron Microscopy, Infra-Red Spectroscopy/. Monomolecular reaction of thermal and electron impact decomposition in solids and gases.
1978-1995 - USSR State Department of Microbiology, Institute of Biological Instruments Design and Construction, Moscow. Design of portable Gas Chromatography - Mass spectrometry (GC-MS) systems. Research and development in GC and GC-MS methodology of detection the micro-organisms and monitoring of microbial communities in environment, medicine and biotechnology by its chemical markers and profiles. Head of GC-MS group.
1995-1998 - Russian Academy of Medical Science, Academician Yu.Isakov Scientific group. Leading Scientist. GC-MS detection of micro-organisms in human body fluids. Monitoring of infection in heart surgery. Detection and monitoring urogenital infections and disbiosis.
1997, October-November - Lund University, Department of Clinical Microbiology. Dr. Lennart Larsson laboratory. Fellowship. Detecting microbial markers in blood of healthy persons and inflamed patients by using GC-MS single ion monitoring technique.
Most important publications:
|
Who’s Who in Microbial
Diagnostic
|
The method was originally developed and introduced by Dr. of Sci. G.Osipov (osipovga@rusmedserv.com) in collaboration with the staff of The State Institution of Biological Instrument Design and Construction (Moscow, Russia), including Dr. A.Dyomina, Dr. T.Nedorezova, Dr. E.Shabanova, junior scientists T.Radiushina, T.Krymtzeva. Analytical procedure and methodology was further improved during 1996-98 by creative effort of Dr. of Sci., Prof. N.Beloborodova (beloborodova@rusmedserv.com), junior scientist N.Boiko, Dr. V.Pozdorovkina, Dr. A.Biryukov (bir_alex@rusmedserv.com) in close cooperation with Bacteriology Department of Filatov Pediatric Hospital (Moscow) with kind support and assistance of Dr. V.Kurchavov (kurchavov@rusmedserv.com), the chief of department.
|
Fundamental Foundation
|
Investigation of microbial fatty acids (FA) and chemodifferentiation of microorganisms over the last 30 years are the roots of Microbial Diagnostic described in this report. They are presented by world-wide known investigators like C.W.Moss, E.Yantzen, D.B.Drucker, C.Asselineau, M.Goodfellow, D.E.Minnikin and also scientist like Z.P.Vasyurenko, L.V.Andreev, V.I.Sedova, E.A.Kiprianova et al. in Russia and former Soviet Union.
The phospholipid fatty acids (PLFA) method was a starting point for the research carried out by the group of D.C. White in University of Tennessee (Knoxville) (milipids@aol.com) and his followers like D.Ringelberg and P.D.Nichols (Peter.Nichols@marine.csiro.au) on microbial community structure in environment. They were the first who used Gas Chromatography – Mass Spectrometry (GC-MS) for this purpose. Our effort is further development of White’s novel approach: in addition to phospholipid signature markers, we consider complete microbial cellular FA profiles for figuring out community composition at the genus and, in some cases, even the species level both in environment and medicine.
Nobody else ever before dealt with detection and quantitative analysis of mixed infection and microbial communities in clinical samples. Single marker method and GC-MS technique was systematically used by Lennart Larsson (lennart.larsson@mmb.lu.se) in clinical practice (Lund University, Lund, Sweden), so we collaborate with his laboratory currently.
|
What “Microbial Diagnostics” is?
|
“Microbial Diagnostics” is diagnosis of infection, inflammation and loss of normal flora in human body by detection specific microbial substances – markers -- especially fatty acids, sterols and other lipid compounds and microbial metabolites using Gas Chromatography - Mass Spectrometry Single Ion Monitoring (GC-MS-SIM) analysis and lipid compounds profile balance. The method allows quantitative determination of genera-species composition of mixed infection and microbial communities in body fluids or environment.
The features are:
Presence of different anaerobic slow growing, non-fermentative bacteria, as well as microscopic fungi, and intracellular parasites (viruses, Chlamidia, Neisseria) in human organs complicates study of inflammation or infections by traditional methods. Gas Chromatography - Mass Spectrometry was used for identification and evaluation of mixed infection by the presence of chemical compound of microbial origin in blood, urine, sputum, pus, vaginal liquid, sperm etc.
This short-term analysis (less than 5 hours) is universal, does not require cultivating and excludes biological test specimens. Local database was created which consist of standard fatty acid profiles of pure cultures the microorganisms of clinical value and algorithm of marker control. This program realized as GCMS research based on lipid biomarker analysis and balance equation methodology for mixed infection calculation using Microsoft Excel Tables.
Analysis is based on determination of trace chemical markers in biological fluid. Occurrence of branched and odd acids, hydroxy acids, metabolized sterols and certain aldehydes in body fluid is chemical evidence of bacteria presence. Microbial cells contain more than 200 fatty acids that differ from human ones. Only 10-20 FA from the list of 200 is typical for one single bacterial genus. Thus, characteristic FA markers and profiles allow to distinguish bacterial genera and species one from another and from human cells.
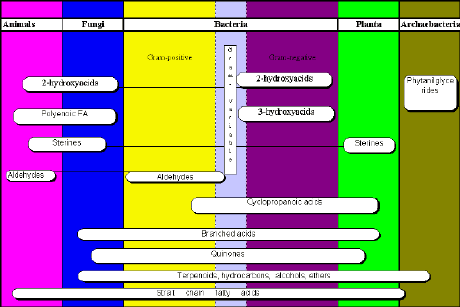
Fig. 1. The diversity of lipid patterns in Nature. Animals, plants and microorganisms of different taxons posses both the same and different lipid components of structural biopolymers
(click on image to see full size)
The dynamic of alterations of fatty acids composition is indicative of their exogenous origin up to the rate 0.001% of total pool of fatty acids in biological fluids.
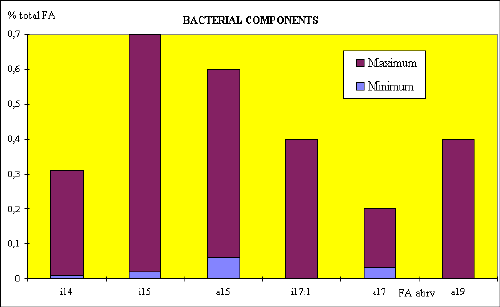
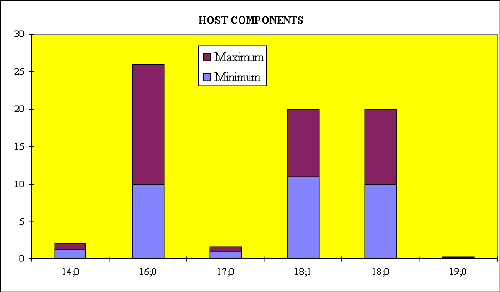
Fig.2. Test for chemical markers
Data processing of more than 500 samples by the ranging method showed the resemblance to the frequency of positive determination of certain microorganisms by culture method as described in literature.
| History and Details |
Human microflora has been shown to contain microorganisms which refer to bacteria, microscopic fungi and protozoa. Some of them are intracellular parasites. Unfortunately, currently available methods are not sufficiently rapid and universal for slow growing bacteria, anaerobes, nonfermenters and other extraordinary microbes.
Chemical methods currently finding increased use to distinguish microorganisms in applied research turn to be far more rapid. The two most widely used methods are gas chromatography (GC) and gas chromatography-mass spectrometry (GC-MS), which are capable of providing extensive information on monomer chemical components of microbial cells and metabolites. Such cellular chemical components (or markers) can be detected among other chemical constituents of any biological object, indicating the presence of the corresponding microbial genera or species in the community under study.
Chemodifferentiation is, of course, widely used for identification of microorganisms isolated in pure cultures. It is based on the use of very large databases containing fatty acids profiles of a few thousand strains of bacteria and microscopic fungi. The Microbial Identification System (MIS), marketed by MIDI Inc., Delaware, is an example of such systems. Fatty acid features are commonly used now in bacterial taxonomy and clinical bacterial diagnostics.
The marker method was a starting point for the research carried out by the group of Prof. D.C.White (University of Tennessee, Knoxville, milipids@aol.com) and his collaborators, who later turned to the analysis of microbial community composition. To quote White, "as certain fatty acids are specific to certain bacteria, it has been the practice to determine microbial community structure by the use of fatty acid markers." What is presented here is an extension of White's approach: we have enlarged the number of markers up to species level. We consider also whole lipid profiles of micro-organisms to calculate community composition at the genera and, in some cases, even the species level.
We have dealt with identification and enumeration of micro-organisms in native samples, and these methods do not require prior growing or isolating colonies on selective media. The local databases we developed for different ecological niches store our own data and those found in literature on the chemical composition of micro-organisms inhabiting this particular niche (or human organs as well). Such a constraint eliminates the ambiguity in species identification inherent to the use of general data banks taking into account that the fatty acids composition of some micro-organisms from different ecosystems may be very close.
Consistent, standardised methods to measure and quantify microbial chemical markers directly in body fluids have not been established. Nevertheless, early, prompt and accurate diagnosis is of importance and prospective for improving strategy of antimicrobial therapy.
Only a few research has been undertaken for clinical diagnostic by using marker method. Well-known is gas chromatographic method for diagnosis of candidacies by arabinitol detection in serum and urine. Muramic acid concentration in blood was used for non-specific detection of bacteria. Other studies used lipopolysaccharide (LPS) specific hydroxy fatty acids for detection of gram-negative bacteria. Moreover, 3-hydroxymyristic acid has been identified as marker of Haemophylus in biological fluids, and used for differentiation of Campylobacter species. Meningococci were detected in blood by measuring 3-hydroxylauric acid content using GC-MS single ion monitoring technique. The same method spotted Neisseria gonorrhoae, and a number of other examples are available.
This approach was broadened to simultaneous determination of more than one bacteria by its lipid markers and profiles as a result of whole biological sample (urine, blood, tissue, pus, etc.) analysis. Fatty acid concentrations were considered in balance equations with partial include of single micro-organisms proposed as community members. Computerised processing appears to be effective in our recent discovery of genera-species composition of some ecological microbial communities. Furthermore, the method have been transformed to detection symbiotic and invasive microflora and identification of microbial markers in body fluids with their 104 background lipid prevalence.
| Comparative Features |
The capacity of classic microbiological approach, i.e. isolation of bacterial strain through cultivating in selected media with further quantitative analysis by serial dilution’s method, is quiet limited. This method is of little use when composition of microbial communities is to be determined. Firstly, because strain isolation in this particular situation will require a lot of time (weeks), consequently acute period processes stay beyond control/informed interference. Secondly, only a very small number of micro-organisms present in clinical or environmental samples (0.1 to 10%) can be accounted for through enumeration of colony-forming units (CFU).
Modern PCR method have only limited application in clinical practice. It could be used for detection of true pathogen such as Neisseria, Chlamidia, Treponema, Aspergillus which did not present in healthy individuals. Meanwhile symbiotic organisms, such as Bacteroides, Candida, Streptomyces, Bacillus, Clostridium, Lactobacillus, Staphylococcus, Streptococcus, and numerous enterobacteria and enterococci are to be always detectable in human specimens, in spite of inflammation being. So PCR is out of significance for detection of inflammation, because of it is only qualitative method but not quantitative one (limited to detection of purely pathogenic bacteria).
GC-MS method is both qualitative and quantitative. When inflammation occur, then microbial marker exceed normal statistical value.
Our method consists in simultaneous measurement of concentrations more than 100 fatty acid and sterol markers of micro-organisms representing the major symbiotic and invasive human microflora. It is performed by using gas chromatography - mass spectrometry single ion monitoring (GC-MS-SIM) technique. The method is express (less than 5 hours per analytical cycle), sensitive (103cells/ml) and universal, irrespective to type of bacteria: anaerobic, aerobic, slow or fast growing, fermenting or not. Microscopic fungi, yeast’s, viruses, protozoa and intracellular parasites could be analysed too.
Specific substances are used as chemical markers for different organisms such as Bacteroides, Fusobacterium, Candida, Streptomyces, Lactobacillus, Staphylococcus, Haemophilus, Neisseria, Chlamidia, Streptococcus, Mycobacterium, Herpes-virus, Treponema, Aspergillus, Helicobacter pylori and so on. Data base does include data on chemical composition of over 1000 strains of 500 clinically valuable species. Computer program was developed including EXCEL calculation for PC. Concentration of single or collective markers, such as hydroxy, cyclopropanoic, unsaturated and branched fatty acids and sterols are used as a measure of bacteria quantity in algorithm combined with balance equations of chemical profiles.
The analysis of lipid moiety by GC-MS-SIM technique provides a convenient way of obtaining quantitative data in spite of surpassing background formed by fluid and epithelial cells components . Known quantity of hydroxy fatty acids or known number of referent bacteria cells were examined in separate experiment for calibration.
| Analytical procedure |
Body fluid (3ml urea; 0,05ml blood, pus, semen or vaginal fluid) was subjected to lipid extraction procedure and acid methanolysis of dry lipid residue in 0,4ml dried HCl in methanol 1N by heating to 800C for 1hr.
Phospholipid FAME (fatty acid methyl esters), LPS hydroxy FAME and dimethylacetales from plasmologen were formed as a result and were extracted with hexane. The hexane fraction were dried, and dry residue was silylated with BSTFA by heating to 800C for 15min.
GC-MS analysis could be performed on a Hewlett-Packard HP-5985B, 5993, 5973 systems or Shimadzu QP-2000-5050 or any modern instrument with a cross-linked methyl silicone capillary column (any like Ultra-1of Hewlett-Packard). The oven temperature was 2 min at 1200 C and then programmed to 3200C at 5 grad/min.
|
Markers in blood and Human Infectious status |
1. Infectious status of human as measured by microbial markers in blood.
| Today the well known sense that one drop of blood contains information about a person’s health could be broadened to understanding that it contains also information about person’s infectious status. State of coexistence of potential pathogens with the host. We must learn to understand the meaning of equilibrium and meaning of deviations from that. |
Prof. Nataliya V.Beloborodova was the first who introduced this approach in everyday clinical use. She supported researches and applications the method in heart surgery, pediatry, septicemia, abdominal and urogenital infection diagnosis and monitoring.

Photo: Prof. N.Beloborodova and G.Osipov
Our investigations of microbe markers in blood present new approach in detection of pathogenic micro-organisms in human body without culturing specimens from various infectious locus’s. We have studied over 200 blood samples from donors and patients up today and the same number from experimental animals. We’ve estimated the whole meanings of such data and it’s possible application in clinical practice.
State of the art is as follows.
We measure concentration of lipid substances in blood which assigned to definite genera, group or species of micro-organisms.
|
Table of markers |
Certain small molecules, found in human blood, are related with maximal probability to definite groups of microorganisms. These data are based on the results of our investigation on chemical composition of bacterial cell fatty acids, as well as on digest of scientific literature on the issue, in particular on chemodifferentiation of bacteria.
Database of chemical substances, related to those microorganisms, in which they are found the most often.
|
¹ |
Code* |
Chemical name |
Characteristic microorganism |
|
1 |
Ñ10 |
decanoic |
Streptococcus |
|
2 |
a13 |
anteiso-tridecanoic |
Bacillus cereus |
|
3 |
i14 |
iso-myristic |
Peptostreptococcus anaerobius, Streptomyces, Bacillus |
|
4 |
14:1D 9 |
9,10-tetradecanoic |
Streptococcus pneumonia, Clostridium |
|
5 |
i15 |
iso-pentadecanoic |
Propionibacterium, Bacteroides, Staphylococcus |
|
6 |
à15 |
anteiso-pentadecanoic |
Staphylococcus, Bacillus, corinebacteria |
|
7 |
i16:0 |
iso-palmitic |
Bacillus, Streptomyces, Bacteroides, Brevibacterium |
|
8 |
i17:1 |
iso-pentadecenoic |
Flavobacterium |
|
9 |
i17:0 |
iso-heptadecanoic |
Bacillus, Prevotella, Propionibacterium |
|
10 |
a17:0 |
anteiso-heptadecanoic |
Corinebacterium (diphteroids) |
|
11 |
17:1 |
heptadecenoic |
Candida albicans |
|
11 |
17cyc |
cyclo-heptadecanoic |
Enterobacteriaceae |
|
12 |
17:0 |
heptadecanoic |
human cells, invariant |
|
13 |
18:1D 11 |
cis-vaccenic |
Serratia, Enterobacteriaceae, Pseudomonas, |
|
14 |
i18 |
iso-octadecanoic |
Bacillus subtilis, Peptostreptococcus, Bifidobacterium, |
|
15 |
19cyc |
cyclo-nonadecanoic |
Enterococcus, Lactobacillus |
|
16 |
a19 |
anteiso-nonadecanoic |
Staphylococcus |
|
17 |
10Me18 |
10-methyl-octadecanoic (tuberculostearic) |
Mycobacterium, Corinebacterium xerosis group, Ñ.urealyticum |
|
hydroxy-acids: |
|||
|
18 |
h10 |
hydroxy-decanoic** |
Pseudomonas |
|
19 |
h12:1 |
hydroxy-dodecanoic |
Pseudomonas aeruginosa |
|
20 |
h12 |
hydroxy-lauric |
Pseudomonas, Neisseria, Vibrio, Acinetobacter |
|
21 |
h13 |
hydroxy-tridecanoic |
E.coli, Bacteroides hypermegas, Selenomonas |
|
22 |
h14 |
hydroxy-myristic |
Enterobacteriaceae, Fusobacterium, Neisseria, Haemophylus, Moraxella, Campylobacter |
|
23 |
hi15 |
hydroxy-iso-pentadecanoic |
Prevotela |
|
24 |
h16 |
hydroxy-palmitic |
Bacteroides urealyticum, Brucella, Wolinella, Fusobacterium, Bordetella, Campylobacter |
|
25 |
hi17 |
hydrohy-iso-heptadecanoic |
Bacteroides fragilis |
|
26 |
h18 |
hydroxy-stearic |
Helicobacter pylori, Francisella, Brucella |
|
27 |
10h18 |
10-hydroxy-stearic |
Clostridium perfringens |
|
28 |
hi20 |
hydroxy-iso-eicosanoic |
Chlamidia trachomatis |
|
29 |
h22 |
3-hydroxy-docosanoic |
Chlamidia trachomatis |
|
30 |
2h12 |
2-hydroxy-lauric |
Pseudomonas aeruginosa, Acinetobacter |
|
31 |
2h14 |
2-hydroxy-myristic |
Klebsiella |
|
32 |
2hi15 |
2-hydroxy-iso-pentadecanoic |
Flavobacterium |
|
33 |
coprostanol (cholestanol) |
Eubacterium |
|
|
34 |
ergosterol |
Candida, Aspergillus |
*- marking: 17:1 - 17- is the number of carbon atoms, the figure after colon denotes the number of double bonds; h - hydroxy-acid; a,i - indicates methyl-branching; cyc - means cyclopropanoic acid. For example, 2hi15 means 2 -hydroxy-iso-pentadecanoic acid.
** - 3-hydroxy-acids - if position of hydroxyl is not indicated.
GC-MS method with special analytical program gives us this opportunity in spite of sound blood background. It’s technical problem, and is solved by choosing appropriate chromatographic column, analytical and sample preparation conditions. Appropriate GC-MS tuning could give desirable sensitivity level. So the problem is solved.
As we understood during numerous discussions, the problem of originating and assigning of analytical results still remains unclear. We should like to divide this problem into separate questions.
First question. Microbial origin of markers.
General considerations and our experience confirm the microbial origin of lipid components which we consider as pathogen markers. Only strictly bacteria-specific substances were used for detection.
More examples could be drawn after thorough study of marker analysis results with further comparison of each sample with clinical data.
Second question. How markers come to blood.
People live in an environment containing a large number of various microbes. Each person has more than 105-6 bacteria on skin surface and more than 1011-12 of his/her own on intestine mucous. To sum up, the number of bacteria cells on skin surface and mucous membranes overpasses the number of host’s own cells - it’s a well known fact. Moreover, if to recall the phenomenon of bacterial translocation from mucous into sterile tissues and blood. As it is hosts cells ,especially phagocytes, obviously get in touch with bacteria all 24 hours daily during the whole life.
Bacterial markers appearance in blood is the result of immune reaction to pathogen. Microbial antigens (whole cells in that number) are adsorbed and digested by phagocytes, and the resulting products (monomer fragments of biopolymers) come to lymph and blood stream. One should keep in mind that lymph stream constantly rinses tissues and mucus of throat, stomach, intestines and other sites of infections income. Lymph washes out microbial cells - completely or partially digested- and, probably, alive bacteria. Through lymph all microbial matter finally gets to blood, where it’s subjected to second attack of blood immune system, i.e. by neutrophils and immunoglobulins. Phagocytosis - by neutrophils and monocytes in bloodstream and macrophages in tissues - brings about accumulation of numerous foreign to host’s organism substances of bacterial origin in blood. These substances could be bacterial cell fragments, products of biopolymers destruction and others. Moreover, some bacterial metabolites invade bloodstream directly from the gut.
For the purposes of marker analysis it is important that:
The last (accumulation) is important for explanation of virtual large quantity of measured microbial markers. But, another way, who measured it before?
Remark. Not necessary to discuss in details the way which markers come to blood. During a hundred year is known and discussed the phenomenon of translocation the micro-organisms between human organs. It really exist, and we can put the details in “black box”:
|
Throat |
® |
|||
|
Stomach |
® |
Autolysis |
||
|
of microbes |
||||
|
Intestine |
® |
|||
|
Time “T” |
“Black box” of translocation |
®
|
Blood |
|
|
Lungs |
® |
|||
|
Phagocytosis |
||||
|
Skin |
® |
|||
|
Bacteremia |
||||
|
Other organs |
® |
Fig.3. The microbes and microbial markers way to blood.
Consequence 1. Detected by marker method microorganisms do not necessarily show up in blood cultures, for they rarely come alive in the bloodstream. They could be detected only when weakened immune system fails to kill them, i.e. in the case of bacteremia.
Consequence 2 . Little use to discuss whether detected microbes were dead or alive: naturally they are usually dead in bloodstream (with the exception of bacteremia) and shortly alive from the moment of invasion and until the attack of immune system cells. This period, T in fig.3, covers the time needed for phagocytosis, transporting and filtration of antigens and microbial chemical patterns to blood. This period lasts approximately between the neutrophils average life-time (6 hours) as a maximum, and the time needed for transportation of chemicals to blood (minimum). The last one is probably an order of several minutes. This is, for example, equivalent to time of drug absorption from stomach to blood. Do you feel the point when alcohol reaches your blood and brain after a glass of wine? So the time of microbe lipid markers appearance in blood after the lysis of cell membrane is minimal.
Third question. Interpretation the marker profiles.
Detection of microbial markers in blood means that relevant microbes are the object of phagocyte attack on mucus or in bloodstream. Consequently the detected microbe was “interpreted” by organism as pathogenic. Concentration of markers reflects the equilibrium number of interaction between phagocyte cells and the microbes of definite genera. Distribution of concentration of markers in healthy individuals (no inflammation) reflects normal resistance of immune system and form normal level of markers concentrations. If one or more species overcome this resistance - inflammation may occurs.
|
Localisation of microbes. Examples |
According to our opinion, microbial markers in blood are originating from various organs of human body. For example, Petostreptococcus anaerobius may be localised in jaw-bone cavities and respiratory abscesses. Neisseria markers are indicative of meningitis in patients with brain pathology or gonorrhoea in urogenital ones. Arising of Klebsiella marker may be caused by pneumonia; enterobacteria, Clostridium markers - by peritonitis fig.4. Elevated concentration of Helicobacter pylori marker could probably prove the increased activity of this microbe in stomach mucous.
The advantage of method was effectively realized at endocarditis. We followed up over thirty microorganisms before, during and after valvectomy by measuring their markers in venous blood. The result was prompt detection of dramatic increase Staphylococcus haemolyticus at rehabilitation period fig.5. Another example demonstrate more possibility non-invasive detection mixed infection of damaged heart valve. Post-operational valve tissue was analyzed by GC-MS-SIM technique, and markers estimated was close to that predicted by blood analysis fig.6
This means that measurement of microbial markers in blood by GC-MS-SIM method allows to estimate through single run the degree of various microbes’ pathogenicity in any organs at given time. The physician is to define which microbe is the most dangerous at the moment and which organ is inflamed. For this purpose blood marker profile could be used alongside with other traditional methods of micro-organisms detection with simultaneous consideration of other clinical data. Accept and introduce this method, we’ll have at our disposal the universal instrument for effective and appropriate treatment of inflammation. According to the opinion of Prof. of Medicine Natalia Beloborodova (Filatov Paediatric Hospital, Moscow, Russia), who successfully used this approach, the method and GC-MS instruments will gain popularity all over the world in the nearest future. Professor’s main clinical task is reasonable strategy and tactics of antibiotic therapy, primarily directed to reaching reasonable equilibrium between microorganisms in their community, and then microbial community with the host, but not at all eradication of untoward microorganisms.